

The cohort was recruited between Oct 22, 2014, and June 29, 2017, and clinical data were collected until March 31, 2018. The primary endpoint, which was assessed in all fetuses, was the detection of diagnostic genetic variants considered to have caused the fetal developmental anomaly. Genetic results related to fetal structural anomaly phenotypes were then validated and reported postnatally. Sequencing results were interpreted with a targeted virtual gene panel for developmental disorders that comprised 1628 genes.
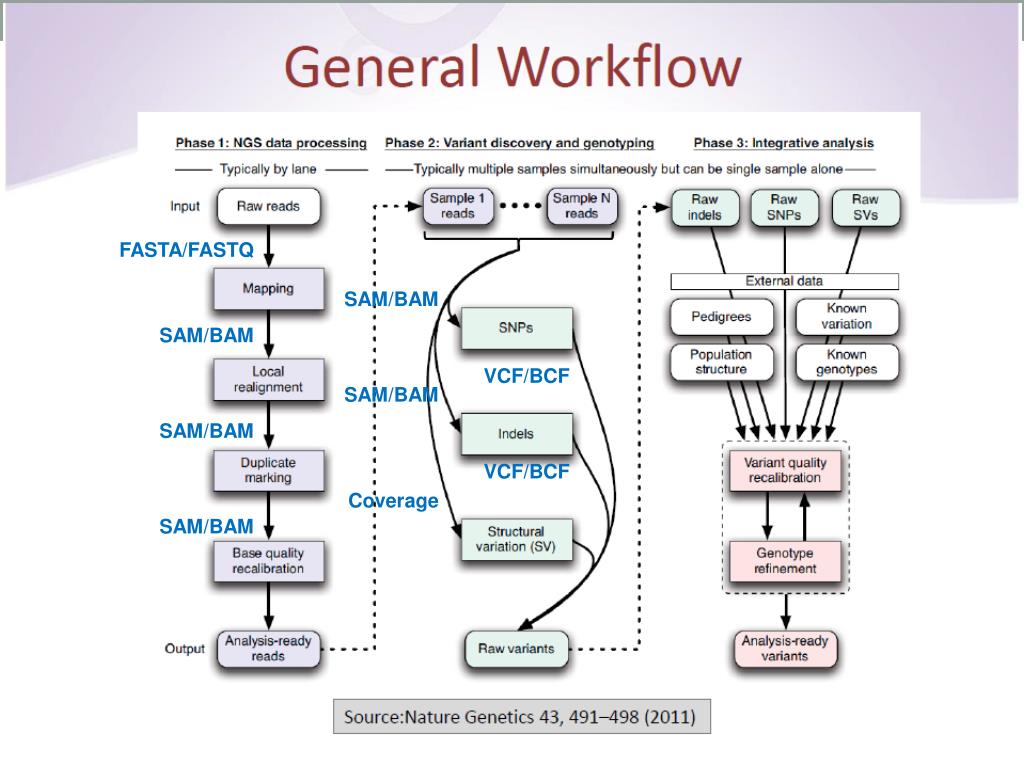
The partners of these women also had to consent to participate. Women were eligible for inclusion if they were undergoing invasive testing for identified nuchal translucency or structural anomalies in their fetus, as detected by ultrasound after 11 weeks of gestation. We used whole-exome sequencing (WES) to evaluate the presence of genetic variants in developmental disorder genes (diagnostic genetic variants) in a cohort of fetuses with structural anomalies and samples from their parents, after exclusion of aneuploidy and large CNVs. In this prospective cohort study, two groups in Birmingham and London recruited patients from 34 fetal medicine units in England and Scotland. The Lancet Regional Health – Western Pacific.The Lancet Regional Health – Southeast Asia.The Lancet Gastroenterology & Hepatology.Genetic variation's effect on high-risk alleleīasic Attributes Class MOLPATH Type Laboratory First Released Version 2.61 Last Updated Version 2.73 Order vs. Genetic variation's effect on drug efficacy Genetic variation's effect on drug metabolism Human reference sequence assembly version Gene mutations tested for in Blood or Tissue by Molecular genetics method Nominalĭescription of ranges of DNA sequences examinedĭiscrete variation analysis overall interpretationĭeletion-duplication overall interpretation Variables that apply to the overall study Master HL7 genetic variant reporting panel Source: Regenstrief LOINC Fully-Specified Name Component Whole exome sequence analysis Property Find Time Pt System Bld/Tiss Scale Doc Method Molgen Additional Names Short Name Whole exome seq analysis Bld/T Display Name Whole exome sequence analysis Molgen Doc (Bld/Tiss) Consumer Name Alpha Whole exome sequence analysis, Blood or tissue specimen Associated Observations Sequencing only the coding regions of the genome is currently less expensive than WGS since less genetic material is sequenced. WES and whole-genome sequencing (WGS) are performed to identify the cause of heritable diseases and for various other applications, including population genetics and cancer studies. Variant types include single nucleotide variants (SNVs), small DNA insertions or deletions (indels), copy number variants (CNVs), or other structural variants (SVs). The exome represents less than 2% of the genome but is estimated to contain about 85% of known disease-related variants. LP248469-1 Whole exome sequence analysis Whole-exome sequencing (WES) is targeted sequence analysis of the protein-coding region of the human genome. 86205-2 Whole exome sequence analysis in Blood or Tissue by Molecular genetics method Active Part Description


 0 kommentar(er)
0 kommentar(er)
